| Japanese top page / English top page |
PAICS
Parallelized ab initio Calculation System based on FMO
|
of prion protein |
|||
|
| |||
| In this page, we give a discription about the intra-molecular interaction analysis of prion protein using the FMO method. The following PPTs were used for the previous presentations. This work has been pablished [1]. | |||
|
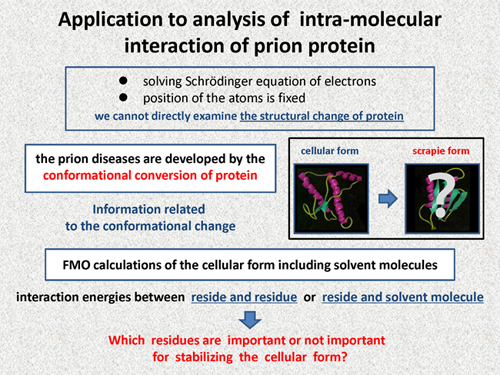
In quantum chemical calculation, we solve the Schrodinger equation of electrons, in which position of the atoms is fixed. Thus, we cannot directly examine the structural change of protein. However, information related to the conformational change is very important because the prion diseases are developed by the conformational conversion of protein, and its path way has been not clear.
In this study, we perform the FMO calculations of the cellular form including solvent molecules. From these calculations, we can obtain the interaction energies between residue and residue or residue and solvent molecule. By analyzing these interaction energies, we try to obtain the followin information about which residues are important or not important for stabilizing the cellular form. We expect that such information will be helpful for the studies of the mechanism of the conformational conversion. |
|||
 |
|||
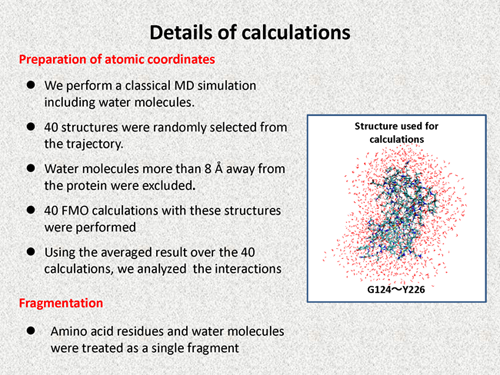
| We perform a classical MD simulation including explicit water molecules. 40 structures were randomly selected from the trajectory, and from which water molecules more than 8 A away from the protein were excluded. 40 FMO calculations with these structures were performed, where amino acid residues and water molecules were treated as a single fragment. An example structure used in our calculations are shown in the follwoing figure. Using the averaged result over the 40 calculations, we analyzed the interaction. | |||
 |
|||
|
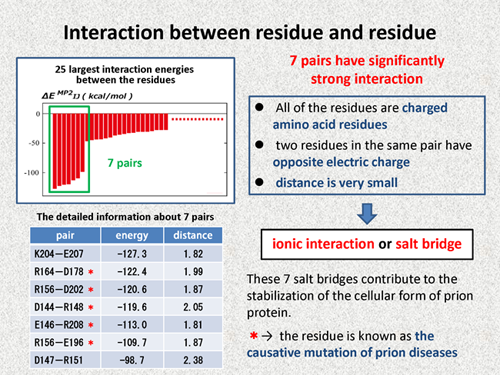
First, I will discuss the interaction between residue and residue. In our calculations, each residue was treated as a single fragment, so we can obtain the interaction energies of all pairs between the residues in prion protein.
This figure shows the 25 largest interaction energies, and we note that 7 pairs have significantly strong interaction. Detailed information about these 7 pairs is summarized in the following table. This table shows that all of the residues are charged amino acid residues, and two residues in same pairs have opposite electric charge, and distance between the two resides is very small. Thus, we can consider that these 7 pairs have ionic interaction or, in other ward, these 7 pairs make salt bridge. These 7 salt bridges contribute to the stability of the cellular form of prion protein. Red marks in the table indicate that the residue is known as a causative mutation of the prion diseases. From this fact, we can consider again that these salt bridges are important for keeping cellar form. |
|||
 |
|||
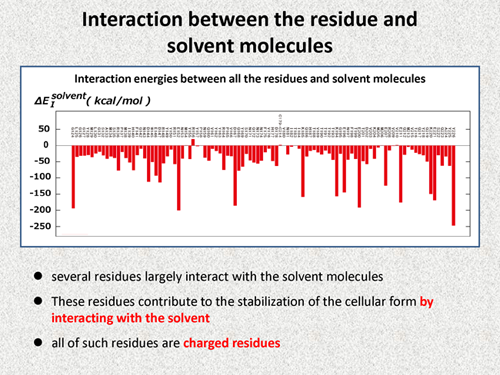
| Next, we discuss the interaction between the residues and solvent molecules. The following figure shows the interaction energies between all the residues of prion protein and solvent molecules. We note that several residues largely interact with the solvent molecule, and we consider that these residues contribute to the stabilization of the cellar form by interacting with the solvent. If we carefully check, we also notice that all of such residues are charged residues. But, this result is not strange because water molecule is very polar. | |||
 |
|||
|
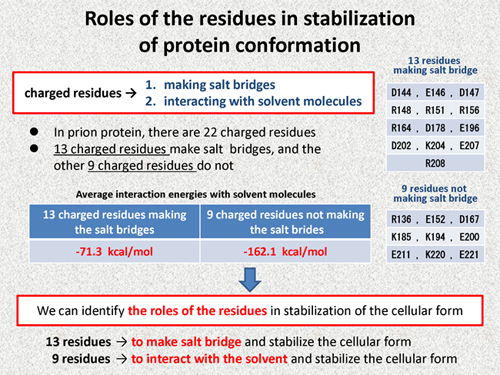
As shown in the above discussions, charged residues contribute to the stabilization of the cellular form by the two ways, that is, making salt bridges and interacting with the solvent molecules.
In prion protein, there are 22 charged amino acid residues, in which 13 charged residues make salt bridges and the other 9 charged residues do not make salt bridge. The following table shows the average interaction energies of these charged residues with solvent molecules. From this table, we note that 13 charged residues making salt bridges have a little interaction energies with solvent molecules, on the other hand, the other 9 charged residues have large interaction energy with solvent molecules. This difference is very clear. That is to say, we can identify the roles of the residues in stabilization of the cellular form: 13 residues have a role to make salt bridge and stabilize the cellular form of protein, on the other hand, the other 9 charged residues have a role to interact with the solvent molecule and stabilize the cellular form of protein. As shown here, we expect that roles of the residues in stabilization of protein conformations can be discussed with the FMO method. |
|||
 |
|||
|
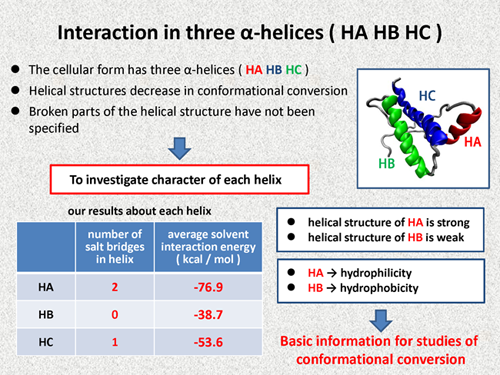
Next, I discuss the interactions in three alpha helices. The cellular form of the prion protein has three α-helices, HA, HB, and HC. And it is known that the helical structures decrease in conformational conversion of prion protein. However, broken parts of the helical structure have not been specified. Of course, only using quantum chemical calculations, it is difficult to specify the broken parts. But, I consider that it is possible to investigate the character of each helix.
In the following table, our results are summarized about each residue. HA has two salt bridges in the helical structure, on the other hand, HB does not have any salt bridge. Thus, in view of the salt bridges, we can consider that helical structure of HA is strong, on the other hand, helical structure of HB is weak. Next, averaged solvent interaction energy of the residues in HA is large, but that of HB is small. Thus, we can say that HA has a hydrophilicity and HB has a hydrophobicity. We consider that these are basic information for studies of conformational conversion. |
|||
 |
|||
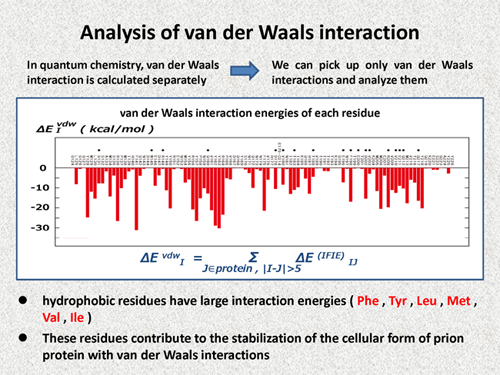
| In quantum chemistry, van der Waals interaction is calculated separately, so we can pick up only van der Waals interactions and analyze them. The following figure shows the van der Waals interaction of each residue. If we carefully check, we note that hydrophobic residues have large interaction energies, e.g., Phy, Tyr, Leu, Met, Val, and Ile. We can realize that these residues contribute to the stabilization of the cellular form of prion protein with van der Waals interactions. | |||
 |
|||
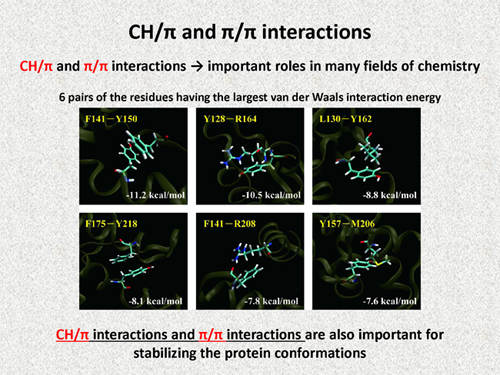
| Recently, CH/π and π/π interactions are considered to have important roles in many fields of chemistry. The folowing figure show the 6 pairs of the residues having the largest van der Waals interaction energy, and all of these are making π/π interactions or CH/π interactions. Thus, our FMO calculations indicate that CH/π interactions and π/π interactions are important for stabilizing the conformation of the native structure of prion protein. | |||
 |
|||
| [1] | Interaction analysis of the native structure of prion protein with quantum chemical calculations, T. Ishikawa and K. Kuwata, J. Chem. Theor. Comput., 6 (2010) 538-547 (DOI: 10.1021/ct900456v) | ||
|